
The weird part is that in my base conda enviornment, _available() returns true.
#CUDATOOLKIT CONDA INSTALL#
I did sudo ubuntu-drivers autoinstall to install the drivers and nothing changed. If I run the install command for nvidia-cuda-toolkit the nvidia-smi command stops working and when I test if cuda is being used with pytorch, I keep getting _available() as False. | 0 N/A N/A 1498 G /usr/bin/gnome-shell 10MiB |īut when I do nvcc -version it returns Command 'nvcc' not found, but can be installed with: sudo apt install nvidia-cuda-toolkit. | GPU GI CI PID Type Process name GPU Memory | | Fan Temp Perf Pwr:Usage/Cap| Memory-Usage | GPU-Util Compute M. | GPU Name Persistence-M| Bus-Id Disp.A | Volatile Uncorr.
#CUDATOOLKIT CONDA DRIVER#
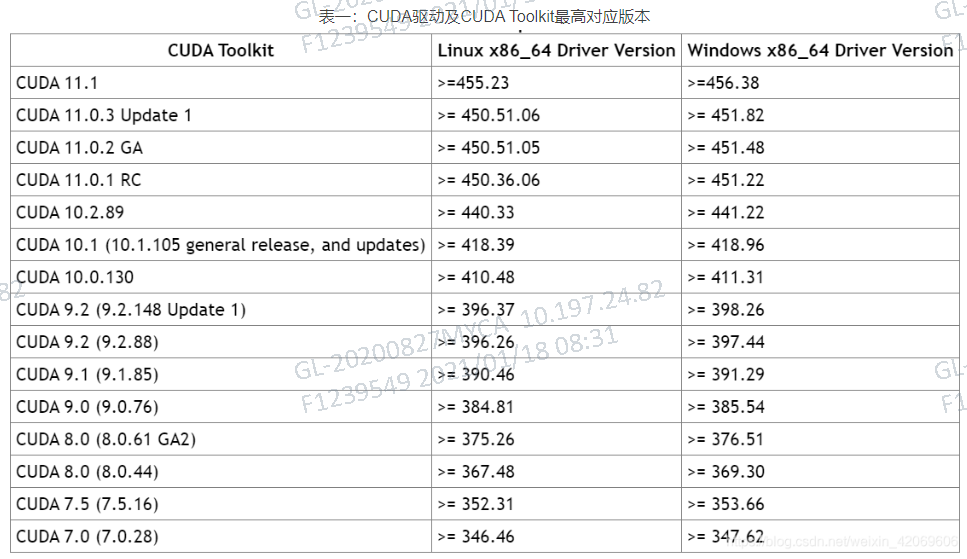
When I enter all the commands I do nvidia-smi and I get this: | NVIDIA-SMI 520.61.05 Driver Version: 520.61.05 CUDA Version: 11.8 | I have Ubuntu 22.04 LTS and I followed these instructions: check the build stringĪ100 GPUs require CUDA 11.I am trying to install Cuda 11.3 (since i understand this is the most stable one and any version after leads to compatibility issues). Try a clean install of Conda and run: conda install pytorch torchvision cudatoolkit, From official site the command is: conda install pytorch torchvision. => WARNING: A newer version of conda exists. (/home/jdoe2/env/a100_env) ~]$ conda install python=3.9 pandas $ module spider ~]$ module load ~]$ conda create -p ~/env/a100_envĬollecting package metadata (current_repodata.json): doneĮnvironment location: /home/jdoe2/env/a100_envĮxecuting transaction: ~]$ source activate /home/jdoe2/env/a100_env/ Note that names that have a trailing (E) are extensions provided by other modules.
#CUDATOOLKIT CONDA HOW TO#
To adopt a modern open data science analytics architecture.įor detailed information about a specific "Anaconda3" package (including how to load the modules) use the module's full name. Srun: job 438605 has been allocated ~]$ module spider anacondaīuilt to complement the rich, open source Python community, the Anaconda platform provides an enterprise-ready data analytics platform that empowers companies You can use the conda search command to see what versions of the NVIDIA CUDA Toolkit are available from the default channels. Srun: job 438605 queued and waiting for resources Install packages into the ~]$ interact -partition=a100_normal_q -nodes=1 -ntasks-per-node=4 -gres=gpu:1 -account=jdoeacct Get an interactive command line shell on a compute nodeĬreate a new anaconda environment at the provided path If you want to use Anaconda on Tinkercliffs a100_normal_q nodes, then you need to build the environment from a shell on those nodes. Instead, use conda list to view and document the most important packages and versions in the environment and build a new environment for the Tinkercliffs normal_q matching those specifications. But you should not use an environment which was built on another cluster (eg. So if you wish to use Anaconda for jobs on normal_q nodes, you can build the environment on the Tinkerliffs login nodes OR on the normal_q nodes. The Tinkercliffs login nodes are essentially identical to normal_q partition nodes. These system differences can make Anaconda virtual environments non-portable between node types.Īs a result, you should create and build a virtual environment on a node of the type where you will use the environment. All are tuned to be customized and efficient for the particular node features. Each node type is equipped with a different cpu micro-architecture, slightly different operating system and/or kernel versions, slightly different system configuration and packages. The Tinkercliffs cluster has at least three different node types and so does Infer.
Create a virtual environment specifically for the type of node where it will be used Instead, use source activate to activate Anaconda virtual environments. # requires the Anaconda initialization from above, and so is not designed to work on systems where a single home directory is shared between several different nodes. "/apps/easybuild/software/tinkercliffs-rome/Anaconda3/2020.11/etc/profile.d/conda.sh"Įxport PATH="/apps/easybuild/software/tinkercliffs-rome/Anaconda3/2020.11/bin:$PATH" _conda_setup="$('/apps/easybuild/software/tinkercliffs-rome/Anaconda3/2020.11/bin/conda' 'shell.bash' 'hook' 2> /dev/null)" # !! Contents within this block are managed by 'conda init' !!


 0 kommentar(er)
0 kommentar(er)
